Download PeptideShaker 2.2.20 (Mac) – Download Free

Download Free PeptideShaker 2.2.20 (Mac) – Download
Peptide Shaker
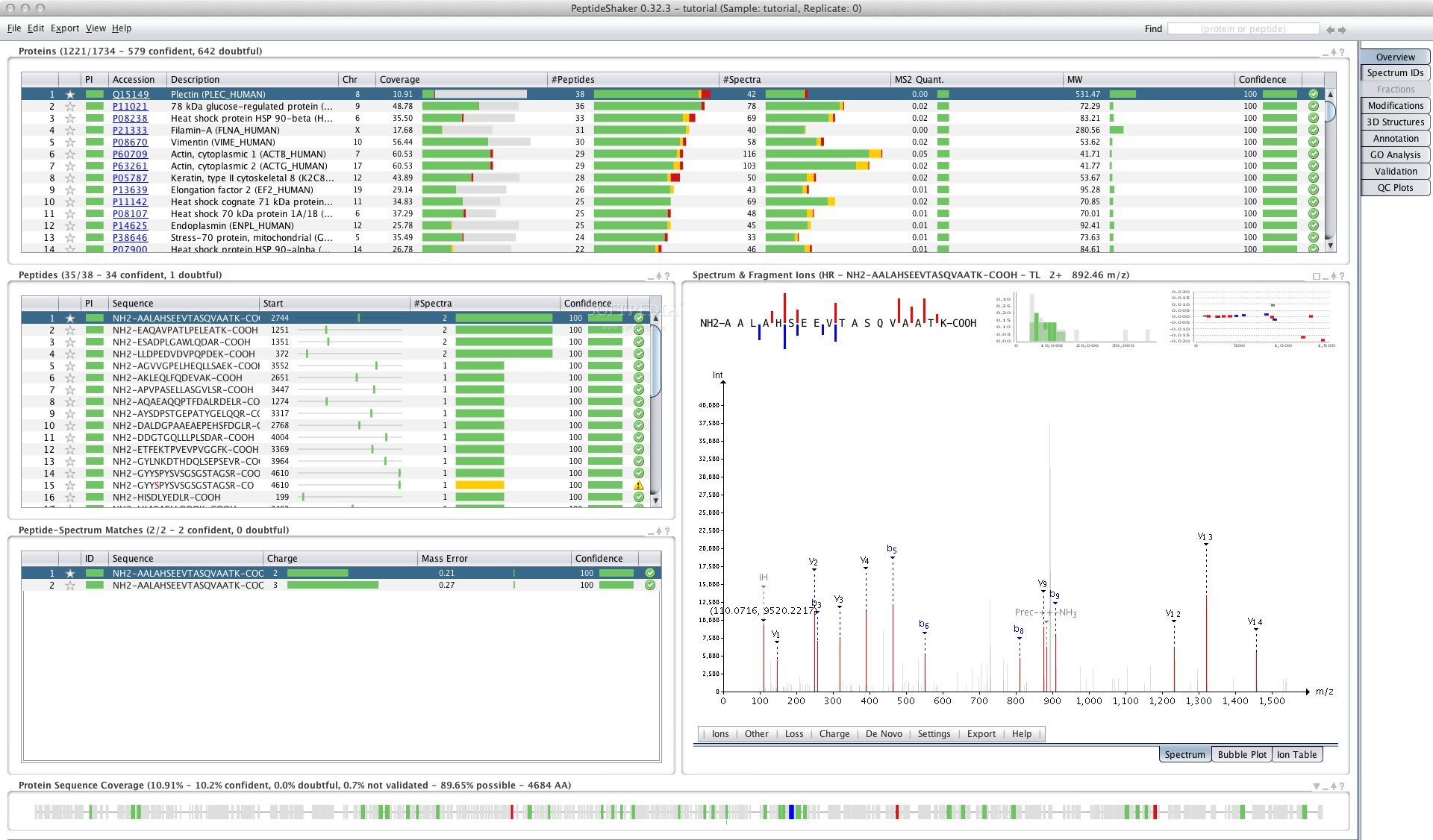
This tool can import results obtained from searchGUI, a GUI for running proteomics identification search engines. Once imported, these results are aggregated into a recognition set. PeptideShaker annotates spectra, calculates consensus scores, handles sequence mapping and protein inference, scores post-translational modification localization, performs statistical validation and quality control, and finally annotates the results using multiple sources of information.
The utility can be used from the command line and allows you to visualize the results using navigation tools. It supports local datasets and datasets stored in the ProteomeXchange PRIDE repository.
PeptideShaker supports nine analysis tasks, allowing you to get an overview of proteins, peptides, and PSMs in your dataset, while also enabling deeper analysis. You can analyze search engine performance and results, examine scores and revisions, draw 3D structure graphs, perform GO augmentation, fine-tune the validation process, analyze results using quality control graphs, and reanalyze public datasets in PRIDE.